Automated assessment of residual plots with computer vision models
ASC2025 talk
Patrick Li
RSFAS, ANU
About me
Hi! I am Patrick Li.
I completed my PhD in Statistics at EBS, Monash University. My research focused on computer vision and data visualization, with an emphasis on developing visual analytics methods to assess residual plots.
I am a postdoctoral researcher at ANU contributing to the Analytics for the Australian Grains Industry (AAGI) project, where my work centres on machine learning, image analytics, and plant phenotyping.
️Co-authors

Prof Dianne Cook
Department of Econometrics and Business Statistics, Melbourne, Monash University

Dr. Emi Tanaka
Research School of Finance, Actuarial Studies and Statistics, Australian National University

Asst Prof Susan VanderPlas
Statistics Department, University of Nebraska-Lincoln

A/Prof Klaus Ackermann
Department of Econometrics and Business Statistics, Melbourne, Monash University, Australia
Challenges in interpreting residual plots reliably
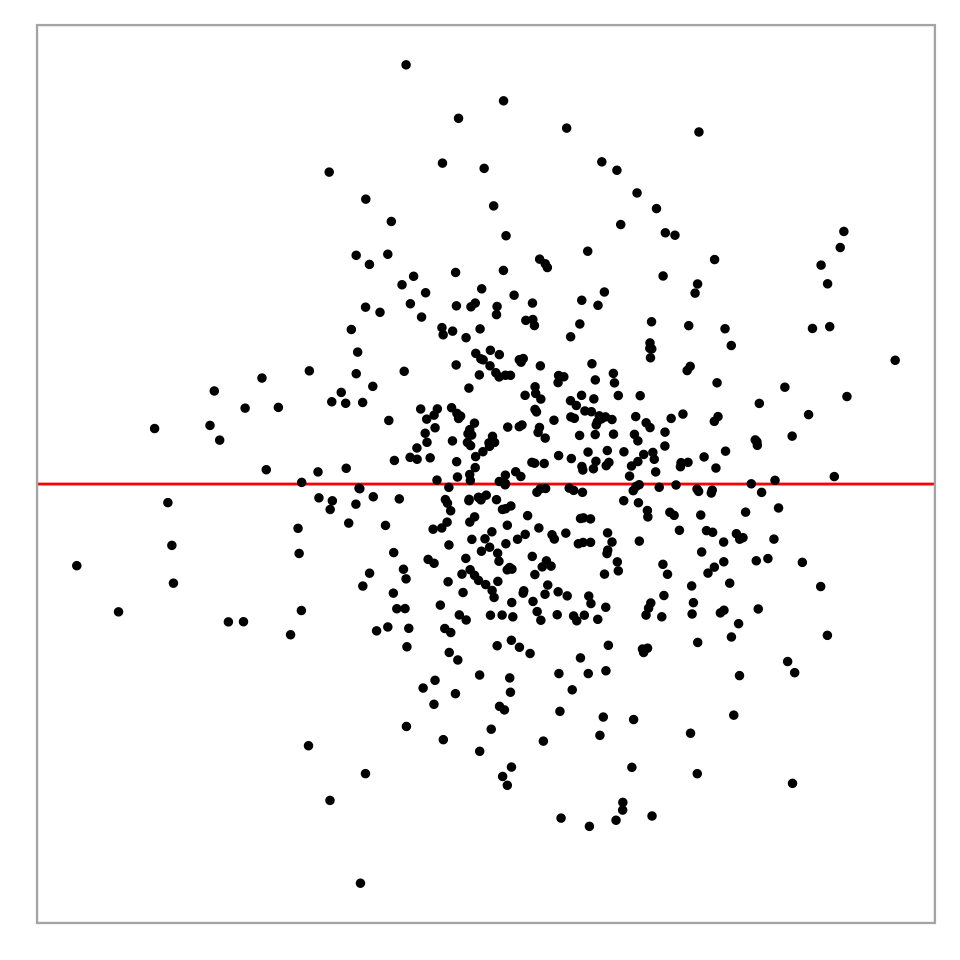
Residual plot of a simple linear regression:
Challenges in interpreting residual plots reliably
Residual plot of a simple linear regression:
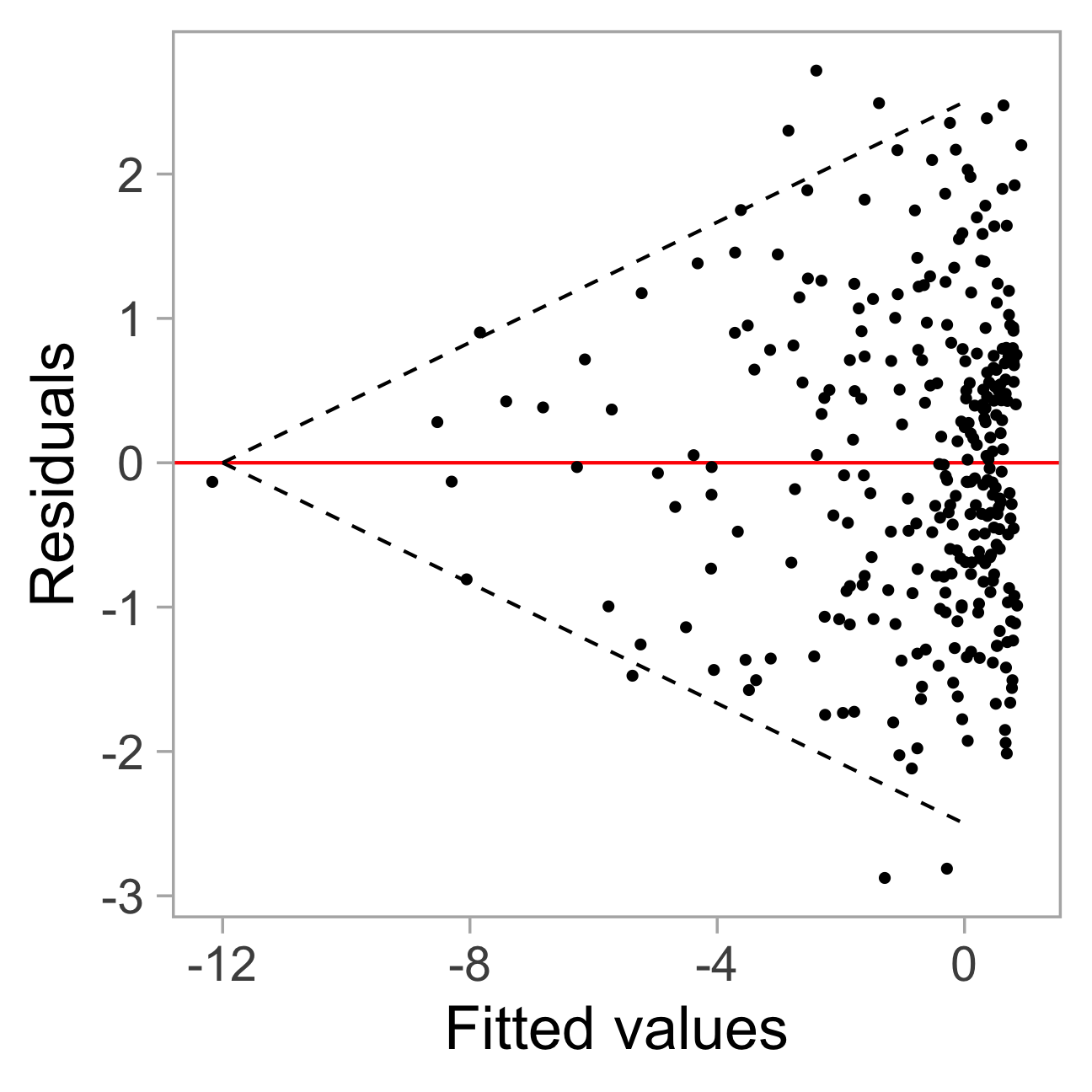
Heteroskedasticity: Vertical spread of the points varies with the fitted values.
However, this is an over-interpretation.
The visual pattern is caused by a skewed distribution of the predictor.
Validity of visual discoveries
Visual discoveries that may appear to violate model assumptions:
- Random variation or sampling noise
- Features of the predictors
- > Genuine model-assumption violations
- …
How do we determine which visual discoveries are valid?
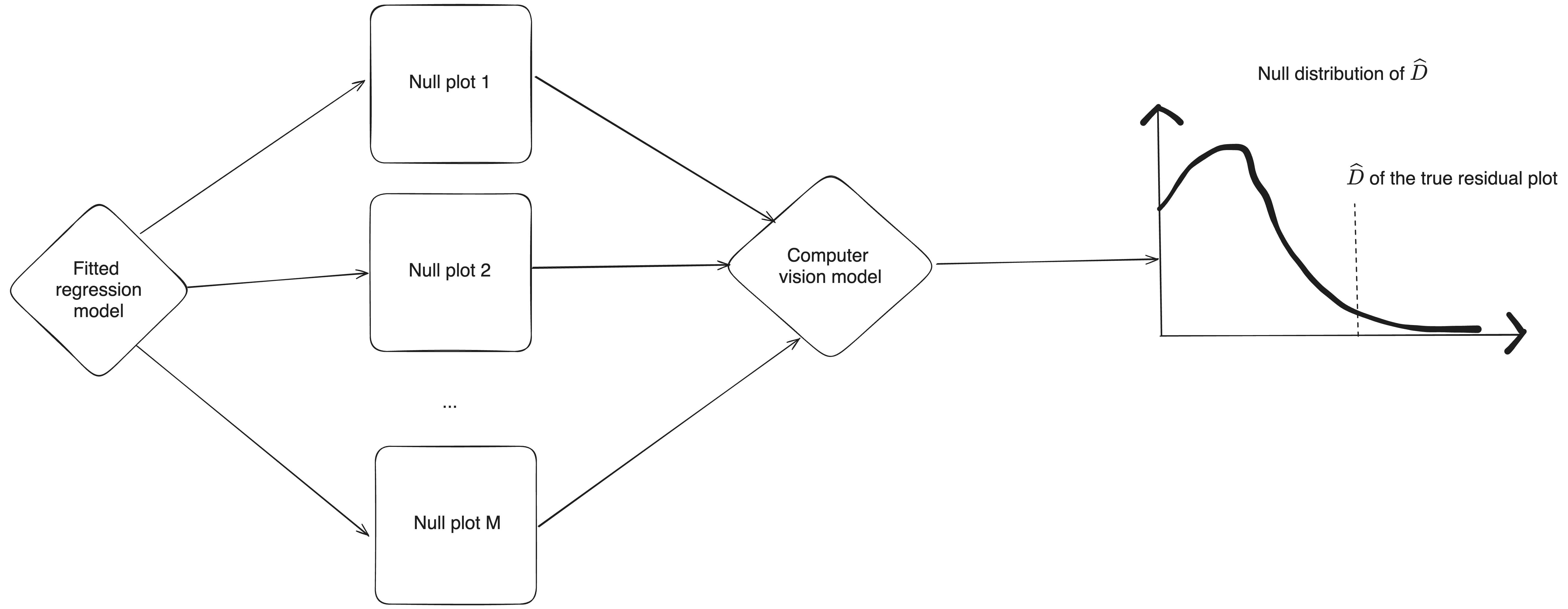
Visual inference (Buja, et al. 2009)
Visual discoveries can be validated by an inferential framework called visual inference.
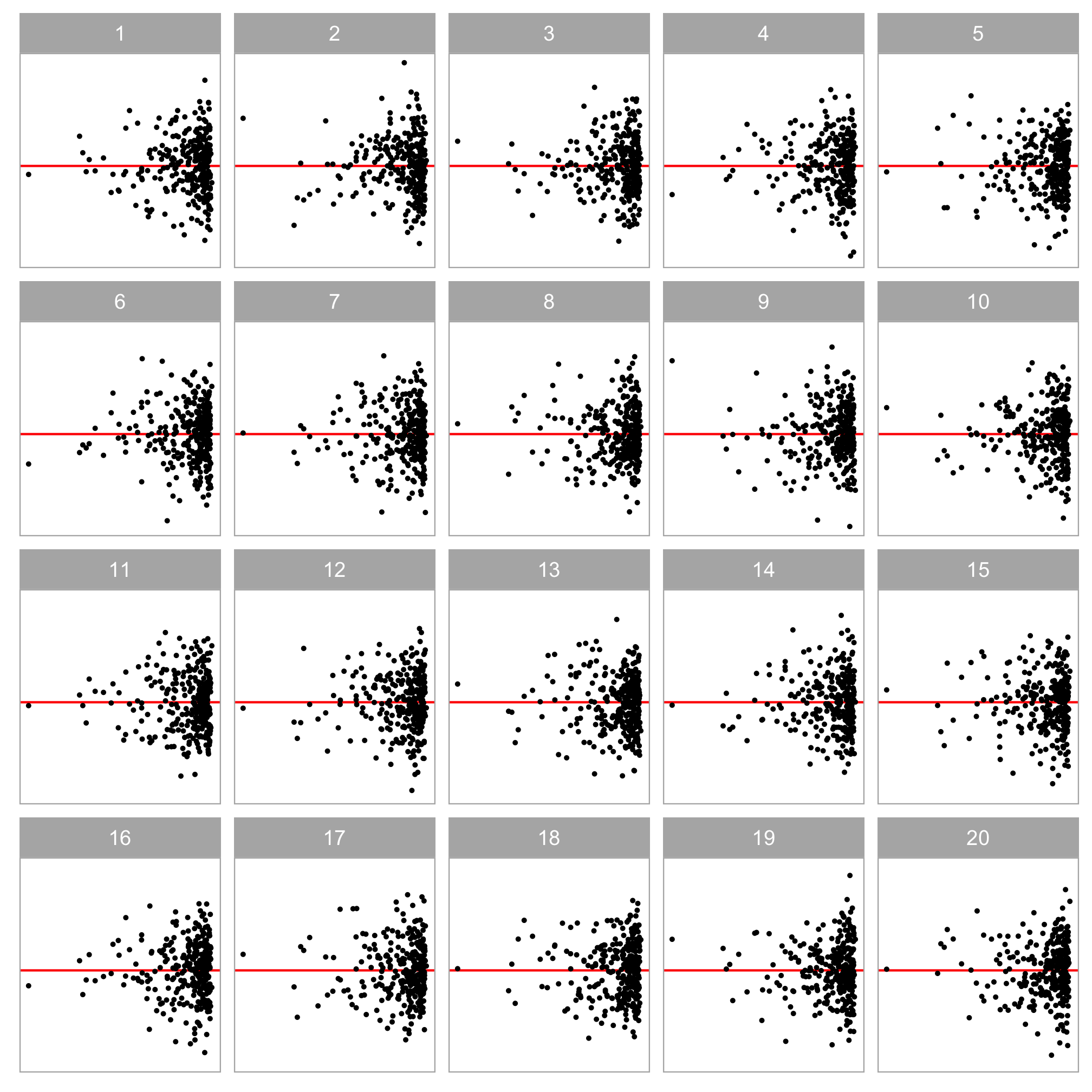
A lineup of residual plots:
- 1 actual residual plot
- 19 null plots containing residuals simulated from the fitted model.
To perform a visual test:
- Ask observer(s) to pick the most different plot(s).
- Calculate the p-value using the beta-binomial model (VanderPlas et al., 2021).
Null residuals simulation (Buja, et al. 2009)
For a classical normal linear regression model:
\hat{\boldsymbol{\beta}} = (\boldsymbol{X}^\top\boldsymbol{X})^{-1}\boldsymbol{X}^\top\boldsymbol{y},\quad \hat{\sigma}^2 = \frac{(\boldsymbol{y} - \boldsymbol{X}\boldsymbol{\beta})'(\boldsymbol{y} - \boldsymbol{X}\boldsymbol{\beta})}{n-p}.
Simulate \tilde{\boldsymbol{e}} \sim N(\boldsymbol{0}, \hat{\sigma}^2\boldsymbol{I})
Obtain \tilde{\boldsymbol{y}} = \boldsymbol{X}\hat{\boldsymbol{\beta}} + \tilde{\boldsymbol{e}}
Estimate \tilde{\boldsymbol{\beta}} = (\boldsymbol{X}^\top\boldsymbol{X})^{-1}\boldsymbol{X}^\top\tilde{\boldsymbol{y}}
Obtain \boldsymbol{e}_{null} = \tilde{\boldsymbol{y}} - \boldsymbol{X}\tilde{\boldsymbol{\beta}}, \quad\hat{\boldsymbol{y}}_{null} = \boldsymbol{X}\tilde{\boldsymbol{\beta}}
Characteristics of the lineup protocol
✅
All-in-one test: can detect many visually recognisable violations
Validated visual findings: helps identify which visual patterns are truly meaningful and may guide model refinement.
❌
Human constraints:
- hard to evaluate many lineups
- hard to process lineups with many plots
Resource-intensive: high labour cost and time-consuming
Can we automate it?
Computer vision model
Modern computer vision models are well-suited for addressing this challenge.
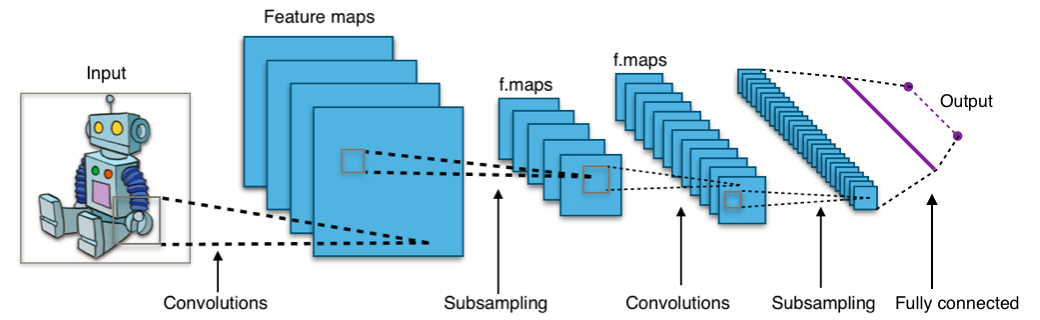
Source: https://en.wikipedia.org/wiki/Convolutional_neural_network
Measure the difference
How do we define a numerical measure of “difference” or “distance” between plots?
- pixel-wise sum of square differences?
- Structural Similarity Index Measure (SSIM)?
- scagnostics?
- …
Distance from a “theoretically” good residual plot
We defined a distance measure based on Kullback-Leibler divergence to quantify the extent of model violations:
D = \log\left(1 + \int_{\mathbb{R}^{n}}\log\frac{p(\boldsymbol{e})}{q(\boldsymbol{e})}p(\boldsymbol{e})d\boldsymbol{e}\right).
P: reference residual distribution assumed under correct model specification.
Q: actual residual distribution.
D = 0 if and only if P \equiv Q.
However, Q is unknown in practice \Rightarrow D can not be computed.
Simulation of training data
In a simulation, we can control the data-generating process and therefore know the true residual distribution Q.
Non-linearity + Heteroskedasticity
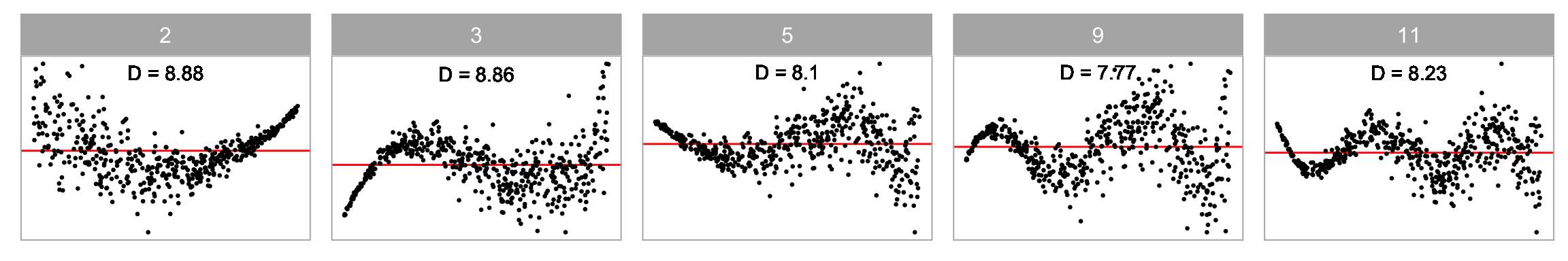
Non-normality + Heteroskedasticity
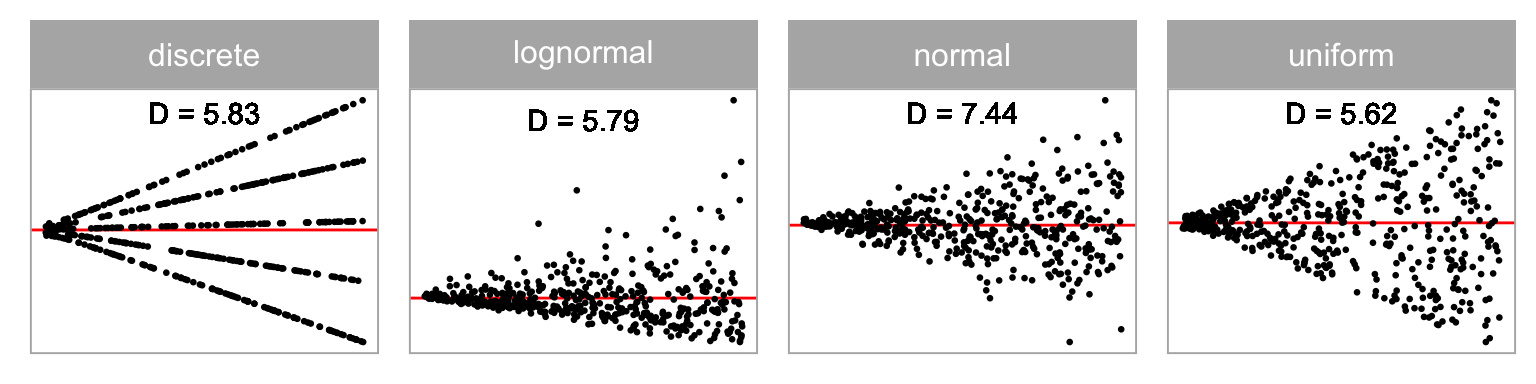
Distribution of predictor

Estimation of D
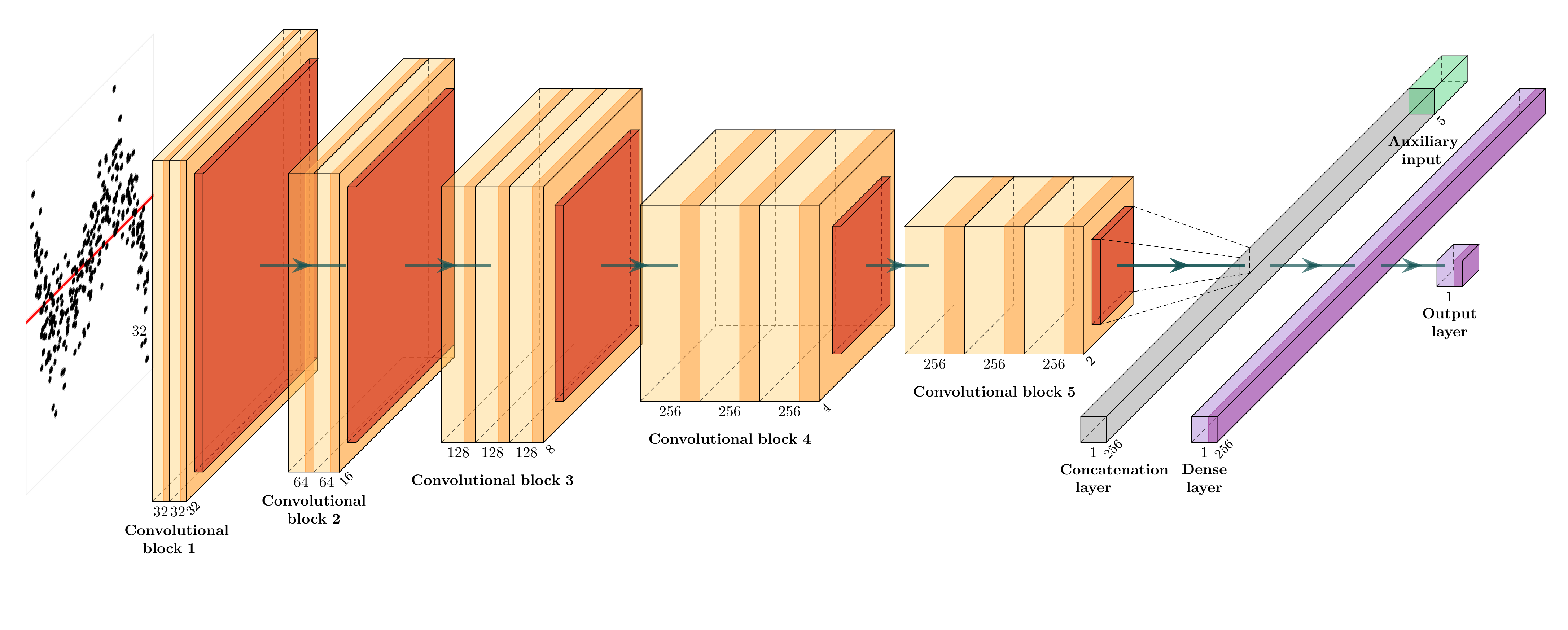
We train a computer vision model to estimate D with 64,000 simulated residual plots:
\widehat{D} = f_{CV}(V_{h \times w}(\boldsymbol{e}, \boldsymbol{\hat{y}})),
where V_{h \times w}(.) generates an h \times w image, and f_{CV}(.) predicts a non-negative distance.
Statistical testing
The p-value is the proportion of null plots having \widehat{D} greater than or equal to the observed one.
autovi Package
Li, W., Cook, D., Tanaka, E., VanderPlas, S., & Ackermann, K. (2025). Automated Residual Plot Assessment With the R Package autovi and the Shiny Application autovi. web. Australian & New Zealand Journal of Statistics.
Core Methods
- Null residuals simulation:
rotate_resid() - Visual signal strength (\widehat{D} prediction):
vss() - Comprehensive checks:
check()andsummary_plot()
Example: Boston housing
library(autovi)
fitted_model <- lm(MEDV ~ RM + LSTAT + PTRATIO, data = housing)
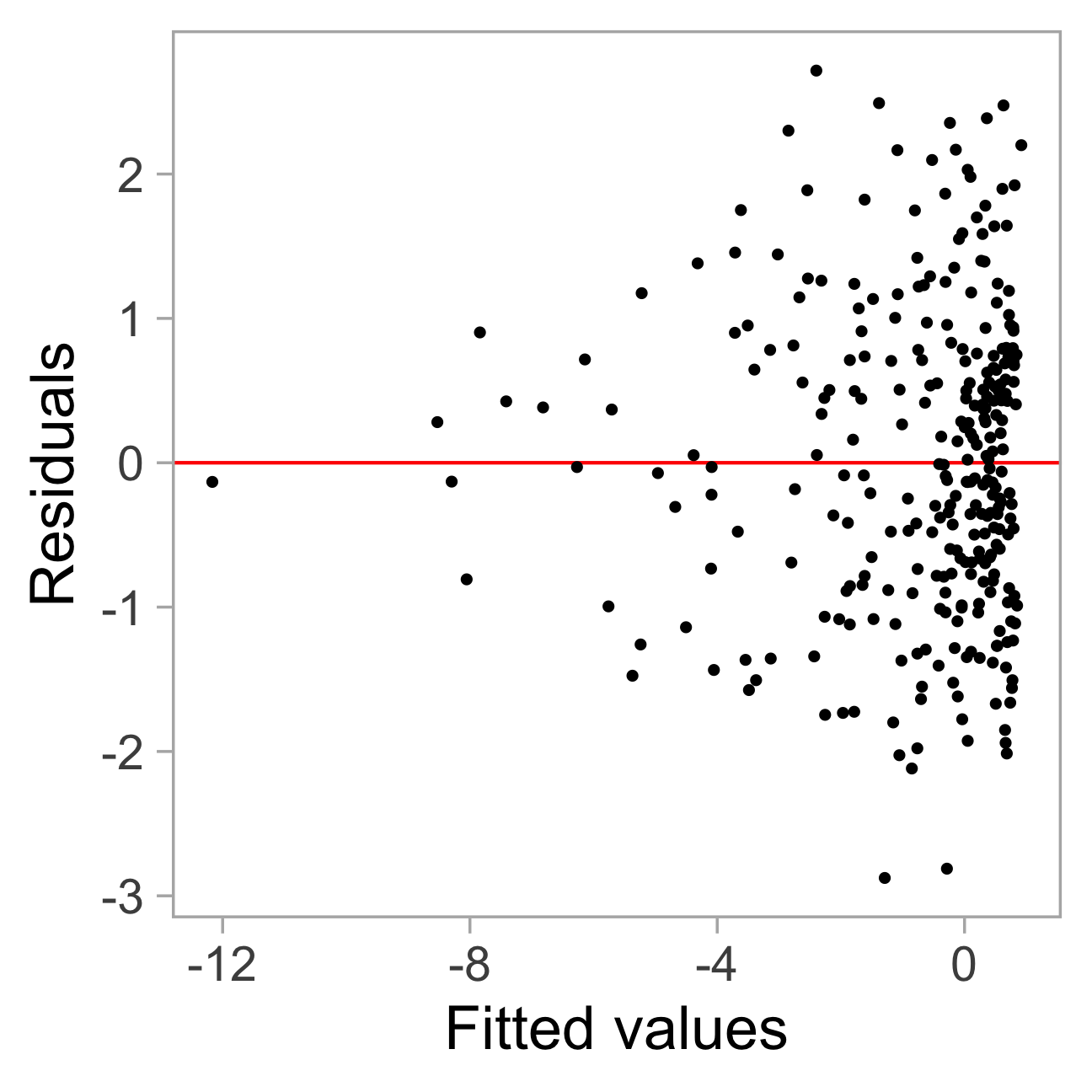
checker <- residual_checker(fitted_model)
checker$plot_resid()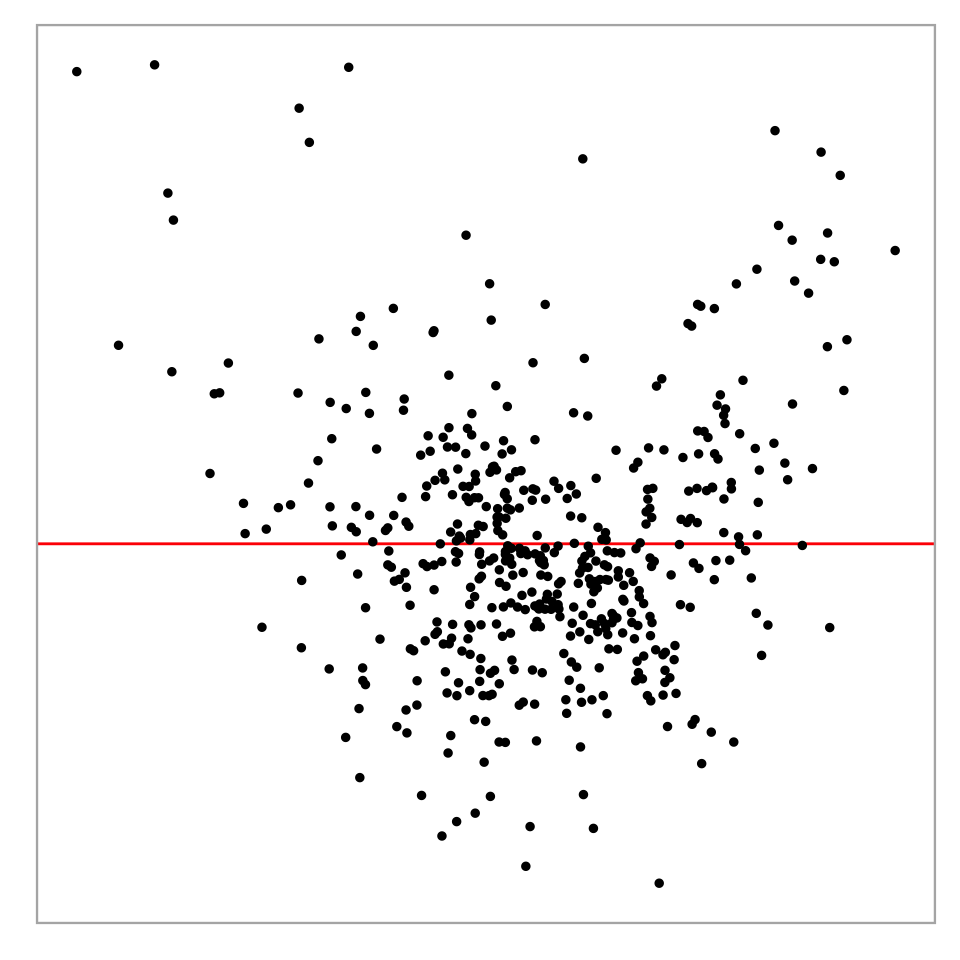
rotate_resid()
Null residuals are simulated from the fitted model assuming it is correctly specified.
# A tibble: 489 × 2
.fitted .resid
<dbl> <dbl>
1 632372. 82404.
2 525177. 24363.
3 646753. -16642.
4 624848. 7895.
5 611817. -25387.
6 551051. -128980.
7 504757. -37748.
8 445700. 33616.
9 281912. -17081.
10 453398. -103580.
# ℹ 479 more rows vss()
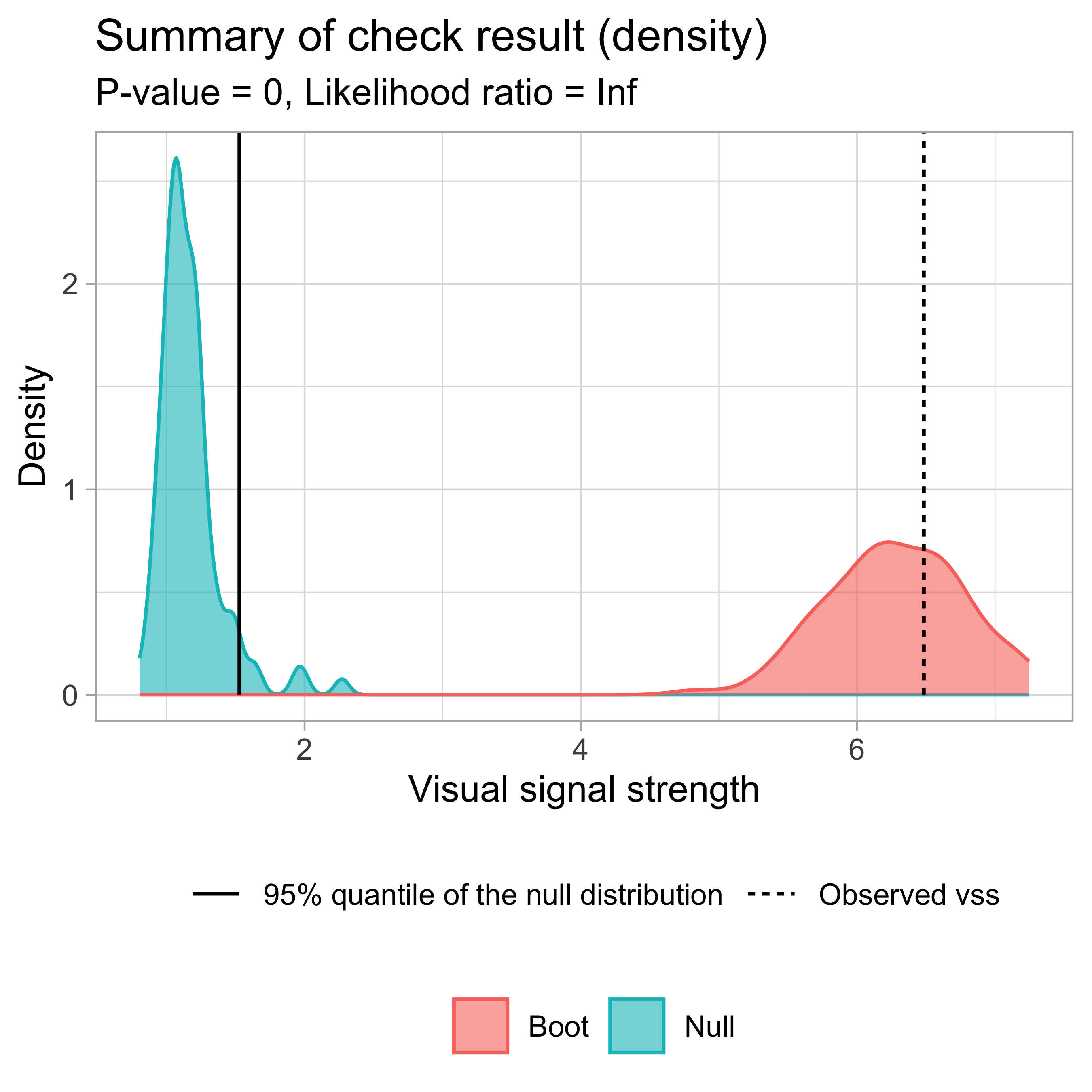
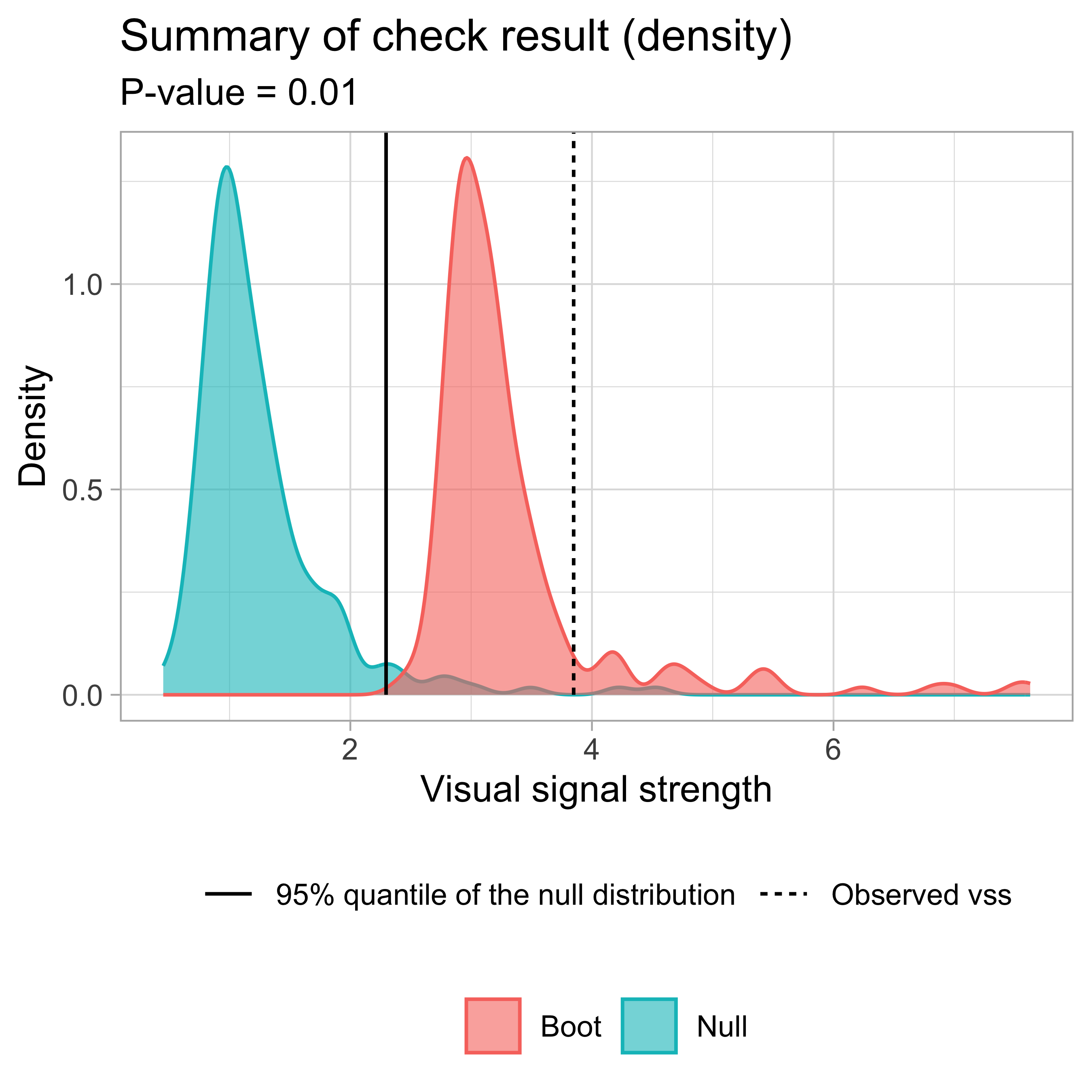
Visual signal strength \widehat{D} of the actual residual plot
✔ Predict visual signal strength for 1 image.# A tibble: 1 × 1
vss
<dbl>
1 6.48 check() and summary_plot()
── <AUTO_VI object>
Status:
- Fitted model: lm
- Keras model: (None, 32, 32, 3) + (None, 5) -> (None, 1)
- Output node index: 1
- Result:
- Observed visual signal strength: 6.484 (p-value = 0)
- Null visual signal strength: [100 draws]
- Mean: 1.169
- Quantiles:
╔══════════════════════════════════════════╗
║ 25% 50% 75% 80% 90% 95% 99% ║
║1.037 1.120 1.231 1.247 1.421 1.528 1.993 ║
╚══════════════════════════════════════════╝
- Bootstrapped visual signal strength: [100 draws]
- Mean: 6.28 (p-value = 0)
- Quantiles:
╔══════════════════════════════════════════╗
║ 25% 50% 75% 80% 90% 95% 99% ║
║5.960 6.267 6.614 6.693 6.891 7.112 7.217 ║
╚══════════════════════════════════════════╝
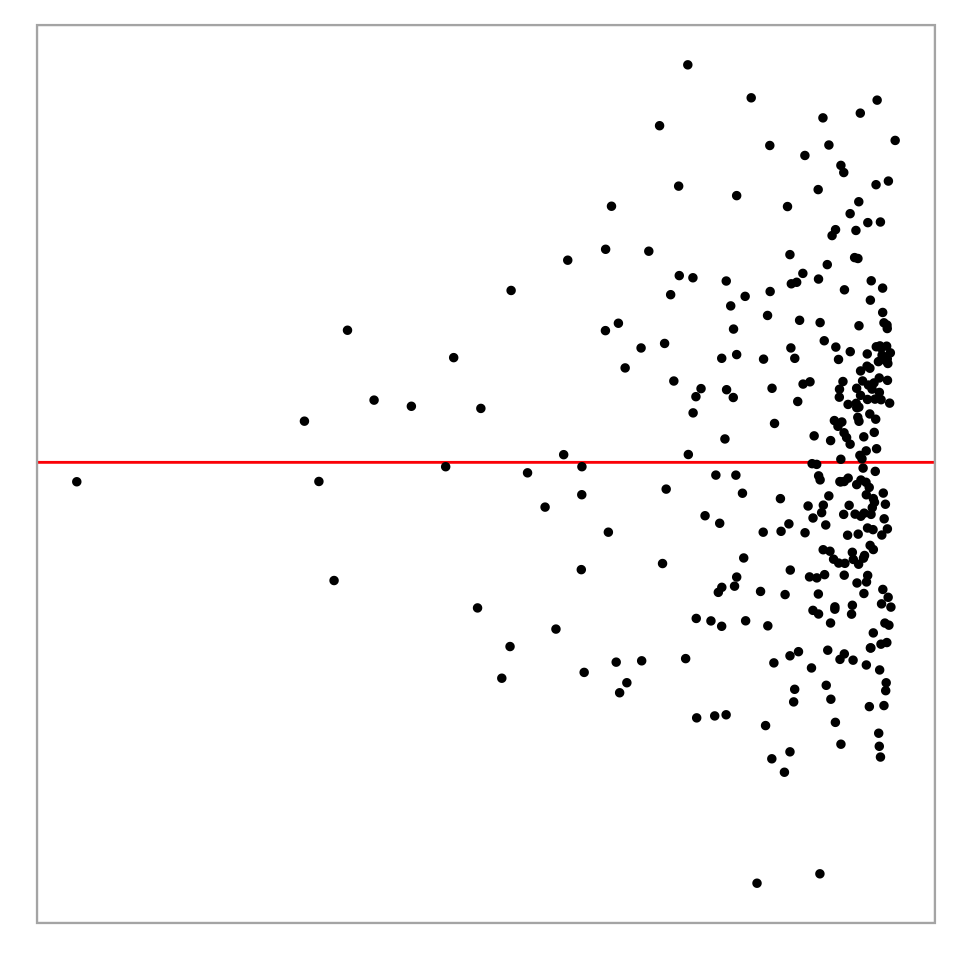
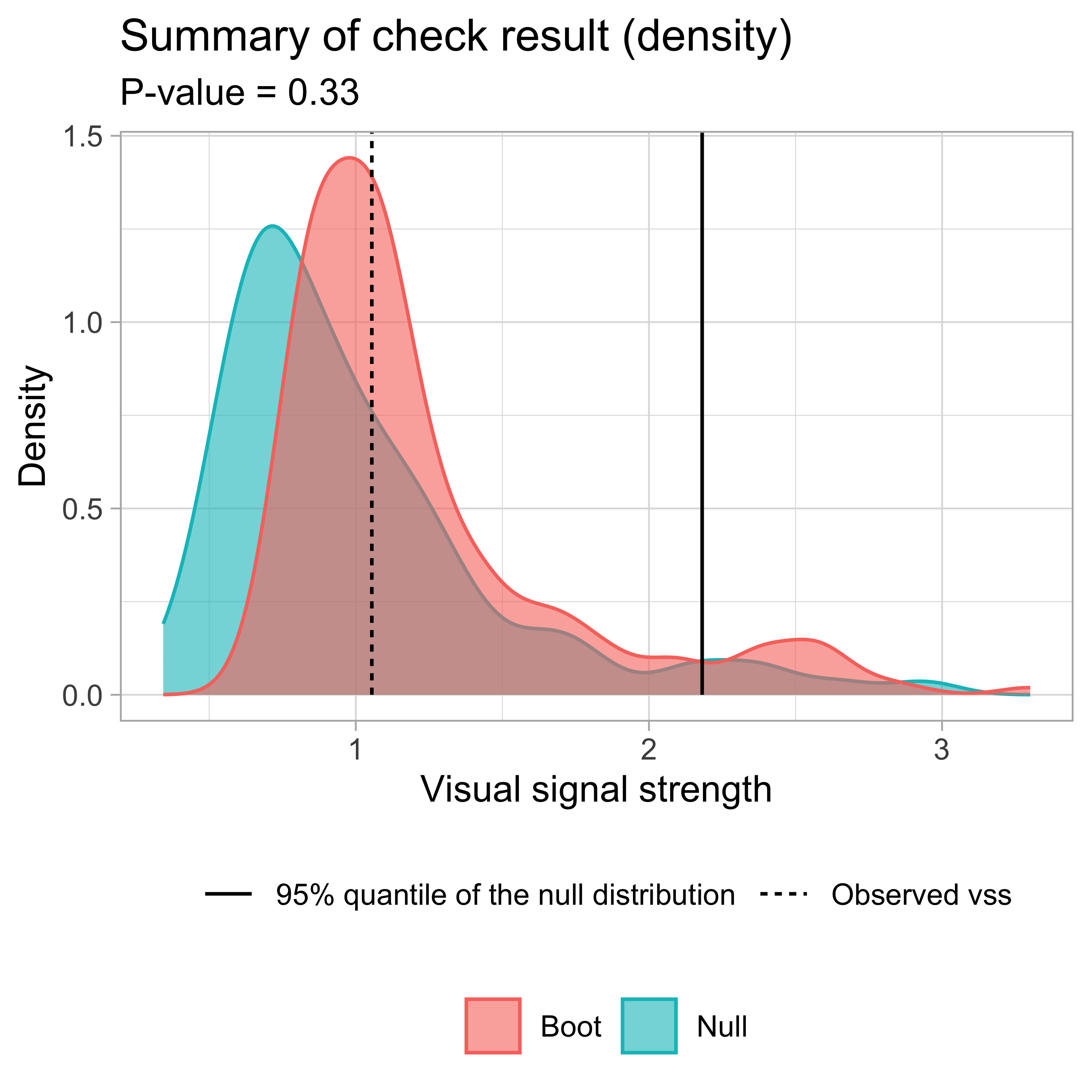
- Likelihood ratio: 0.7064 (boot) / 0 (null) = Extremely large Example: Left-triangle
Breusch–Pagan test p-value = 0.0457
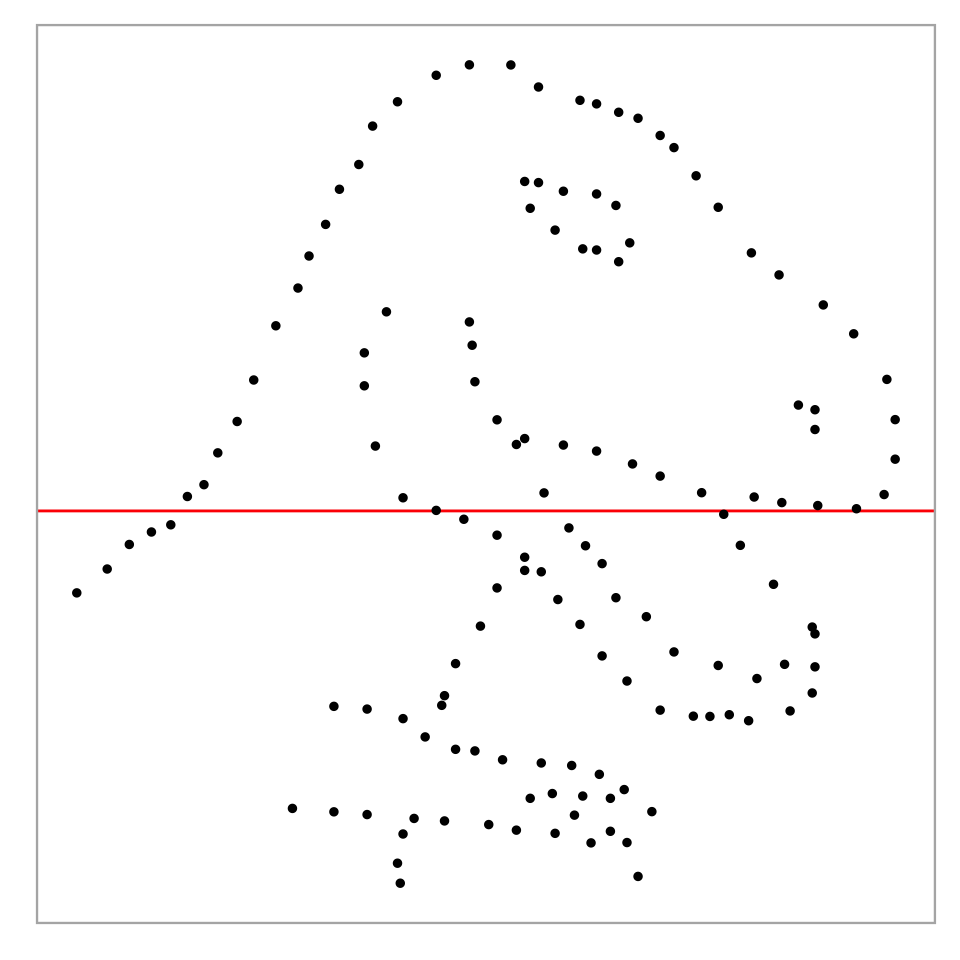
💡Example: Dinosaur
Ramsey Regression Equation Specification Error test p-value = 0.742
Breusch–Pagan test p-value = 0.36
Shapiro-Wilk test p-value = 9.21e-05
🌐Shiny Application
Don’t want to install TensorFlow?
Try our shiny web application: http://autovi.patrickli.org/
Takeaway
You can use autovi to
Evaluate lineups of residual plots of linear regression models
Captures the extent of model violations through visual signal strength
Automatically detect model misspecification using a visual test
Research on extensions to GLM and LMM frameworks is still in progress.
Thanks! Any questions?
Li, W., Cook, D., Tanaka, E., VanderPlas, S., & Ackermann, K. (2024). Automated Assessment of Residual Plots with Computer Vision Models. arXiv preprint arXiv:2411.01001.
tengmcing

Slides URL: https://asc2025-autovi.patrickli.org/ | Perth time